Transgene design
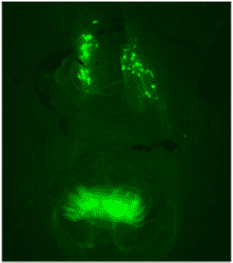 GnRH1:EGFP transgene labels neurons that ultimately control levels of the gonadal hormones testosterone, estrogen, and progestin.
GnRH1:EGFP transgene labels neurons that ultimately control levels of the gonadal hormones testosterone, estrogen, and progestin.
The most efficient way to generate transgenic fish is using the Tol2 system, developed by Koichi Kawakami. This system utilizes elements of the Tol2 transposon from Medaka: the Tol2-encoded transposase is provided in trans with a transgene that is flanked by Tol2 recognition sites. The Tol2 enzyme then catalyzes the insertion of the transgene into the genome.
For a typical vector that overexpresses a transgene in a defined cell population, design a plasmid that contains:
Inject the following into fertilized eggs:
For a typical vector that overexpresses a transgene in a defined cell population, design a plasmid that contains:
- 5' flanking DNA (enhancers/promoter, 5' non-coding exons up through start codon. More 5' region is desirable.)
- Transgene with start codon fused to endogenous start codon
- 3' untranslated region (700-1000 bp of sequence that follows the endogenous stop codon, this is likely to contain miRNA binding site(s) that regulate mRNA distribution.
Inject the following into fertilized eggs:
- Tol2-flanked transgene (225 ng/μL stock; final concentration in injection mix, 75 ng/µL)
- Tol2 mRNA transcribed from pCS-TP (225 ng/μL; final concentration, 75 ng/µL)
- Texas Red-conjugated dextran* (final concentration, 0.5%; 3000 MW; Sigma) for visualization of injection. You can also use 0.05% phenol red, but TxRedDextran will enable you to see whether the injected solution is being taken up into the developing embryo, using a fluorescent dissecting scope.
Transgenic line genotyping
Extract genomic DNA
Take finclip from fish at ≥4 weeks of age into PCR tube.
Add 180 µl of NaOH (50 mM) to each tube.
Heat tubes in PCR machine with FINDNA program: 95ºC 15 min, 4ºC hold.
Add 20 µl of Tris (1 M, pH 8.0) to each tube, mix. Keep tubes at room temp, or freeze for long-term storage.
Amplify
11.8 µl Water
3.0 µl Loading Buffer
2.0 µl Std Pol Buffer
0.4 µl Rev primer (10µM)
0.4 µl Fwd primer (10µM)
0.2 µl dNTP (10 mM)
0.2 µl Taq (NEB)
2.0 µl Template
20 µl TOTAL
Amplify using GENOTYP program:
94º 2:00
94º 0:15 |
62º 0:15 | Repeat 35x
72º 0:45 |
72º 2:00
4º Hold
Detect amplicons
Run on 2% gel to confirm successful amplification: ~45 minutes at 105 V. Expect band at ~300-500 bp, depending on primer pair.
Take finclip from fish at ≥4 weeks of age into PCR tube.
Add 180 µl of NaOH (50 mM) to each tube.
Heat tubes in PCR machine with FINDNA program: 95ºC 15 min, 4ºC hold.
Add 20 µl of Tris (1 M, pH 8.0) to each tube, mix. Keep tubes at room temp, or freeze for long-term storage.
Amplify
11.8 µl Water
3.0 µl Loading Buffer
2.0 µl Std Pol Buffer
0.4 µl Rev primer (10µM)
0.4 µl Fwd primer (10µM)
0.2 µl dNTP (10 mM)
0.2 µl Taq (NEB)
2.0 µl Template
20 µl TOTAL
Amplify using GENOTYP program:
94º 2:00
94º 0:15 |
62º 0:15 | Repeat 35x
72º 0:45 |
72º 2:00
4º Hold
Detect amplicons
Run on 2% gel to confirm successful amplification: ~45 minutes at 105 V. Expect band at ~300-500 bp, depending on primer pair.
Tol2 transposase mRNA synthesis from pCS-TP plasmid
1. Digest overnight DNA with Not l to linearize:
2 µL enzyme
around 5-8ug DNA (high DNA concentration is important)
40 µL total
2. Clean DNA with a spin column
Elute in 8 µL ultra ddH20
3. Take 1uL DNA and run on gel to check if it's completely digested (should give a single, linearized band.
4. Use Ambion mMessage machine Kit:
2 uL enzyme (Sp6)
10 uL NTP/CAP
2 uL Buffer
6 uL DNA (all)
incubate 37ºC for 2 hours
5. Add 1 uL DNase and incubate 37 degrees for 15 minutes
6. Clean up RNA with Qiagen RNA Kit
elute in 50uL
7. Run gel to check if RNA is present:
1uL RNA (incubate prior 80 degrees for 10 minutes)
1.5uL dH20
2.5uL loading buffer that comes with kit
8. Store RNA in -80C
--Thanks to Tod Thiele (U Toronto) & Herwig Baier (MPI Neurobiologie) for the protocol.
2 µL enzyme
around 5-8ug DNA (high DNA concentration is important)
40 µL total
2. Clean DNA with a spin column
Elute in 8 µL ultra ddH20
3. Take 1uL DNA and run on gel to check if it's completely digested (should give a single, linearized band.
4. Use Ambion mMessage machine Kit:
2 uL enzyme (Sp6)
10 uL NTP/CAP
2 uL Buffer
6 uL DNA (all)
incubate 37ºC for 2 hours
5. Add 1 uL DNase and incubate 37 degrees for 15 minutes
6. Clean up RNA with Qiagen RNA Kit
elute in 50uL
7. Run gel to check if RNA is present:
1uL RNA (incubate prior 80 degrees for 10 minutes)
1.5uL dH20
2.5uL loading buffer that comes with kit
8. Store RNA in -80C
--Thanks to Tod Thiele (U Toronto) & Herwig Baier (MPI Neurobiologie) for the protocol.
Tol2 excision assay
Based on Kotani et al., to determine whether the Tol2 enzyme excised the cassette from the vector (and presumably integrated it into the genome).
I haven’t personally needed to do this, but if the fish aren’t carrying the transgene, this may be worth doing.
Ten hours after microinjection, the injected embryos are transferred into 8-strip tubes (we do ~ 15 embryos). After removing water from each tube, each embryo is soaked in 50 ul of a solution containing 10 mM Tris–HCl (pH7.5), 10 mM EDTA and 200ug/ml proteinase K, incubated for 3h to overnight at 50 °C, and heated for 3 min at 95 °C to inactivate proteinase K, 1ul of the DNA sample is used for PCR using primers TYR1 (5'GCT ACT ACA TGG TGC CAT TCC T-3') and BS1 (5'-AAC AAA AGC TGG AGC TCC ACC G-3'), which are designed to hybridize with the plasmid sequence adjacent to Tol2 (Fig. 2). 35 cycles of PCR is carried out: 94°C for 20s; 55°C for 20s; 72°C for 20s, and the PCR products are analyzed on a 2% TAE-agarose gel (Fig. 2). A DNA band of approximately 250 bp can be detected when Tol2 is excised from the plasmid by the transposase activity produced from the coinjected mRNA. Such a DNA band is never detected when the plasmid is injected without the transposase mRNA (Fig. 2). (they toss the injections of there if they less than 80% of the samples have bands).
Reference:
Tomoya Kotani; Saori Nagayoshi; Akihiro Urasaki; Koichi Kawakami. Transposon-mediated gene trapping in zebrafish. Methods (San Diego, Calif.). 2006 , DOI: 10.1016/j.ymeth.2005.12.006, PMID: 16814563
I haven’t personally needed to do this, but if the fish aren’t carrying the transgene, this may be worth doing.
Ten hours after microinjection, the injected embryos are transferred into 8-strip tubes (we do ~ 15 embryos). After removing water from each tube, each embryo is soaked in 50 ul of a solution containing 10 mM Tris–HCl (pH7.5), 10 mM EDTA and 200ug/ml proteinase K, incubated for 3h to overnight at 50 °C, and heated for 3 min at 95 °C to inactivate proteinase K, 1ul of the DNA sample is used for PCR using primers TYR1 (5'GCT ACT ACA TGG TGC CAT TCC T-3') and BS1 (5'-AAC AAA AGC TGG AGC TCC ACC G-3'), which are designed to hybridize with the plasmid sequence adjacent to Tol2 (Fig. 2). 35 cycles of PCR is carried out: 94°C for 20s; 55°C for 20s; 72°C for 20s, and the PCR products are analyzed on a 2% TAE-agarose gel (Fig. 2). A DNA band of approximately 250 bp can be detected when Tol2 is excised from the plasmid by the transposase activity produced from the coinjected mRNA. Such a DNA band is never detected when the plasmid is injected without the transposase mRNA (Fig. 2). (they toss the injections of there if they less than 80% of the samples have bands).
Reference:
Tomoya Kotani; Saori Nagayoshi; Akihiro Urasaki; Koichi Kawakami. Transposon-mediated gene trapping in zebrafish. Methods (San Diego, Calif.). 2006 , DOI: 10.1016/j.ymeth.2005.12.006, PMID: 16814563
